Lim Lab(RNA & epi GENOMICS)

update: 2025.03.05
Selected Papers

1. Gou, L.T.*, Lim, D.H.*, Ma, W.B.*, Aubol, B.E., Hao, Y.J., Wang, X., Zhao, J., Liang, Z.Y., Shao, C.W., Zhang, X., Meng, F., Li, H.R., Zhang, X.R., Xu, R.M., Li, D.S., Rosenfeld, M.G., Mellon, P.L., Adams, J.A., Liu, M.F., and Fu, X.D. (2020) Initiation of Parental Genome Reprogramming in Fertilized Oocyte by Splicing Kinase SRPK1-Catalyzed Protamine Phosphorylation. (*co-first authors)
Cell 180(6), 1212-1227 PMID: 3216915
2. Na, D.K*, Lim, D.H.*, Hong, J.S., Lee, H.M., Cho, D.A., Yu, M.S., Shaker, B., Ren, J.,
Lee, B.M., Song, J.G., Oh, Y.N., Lee, K.G., Oh, K.S., Lee, M.Y., Choi, M.S., Choi, H.S.,
Kim, Y.H., Bui, J.M., Leem K.S., Kim, H.W., Lee, Y.S.#, and Gsponer J.# (2023)
A multi-layered network model identifies Akt1 as a common modulator of neurodegeneration.
(*co-first authors)
Molecular Systems Biology e11801 PMID: 37984409

3. Chen, J.Y.*, Lim, D.H.*, Chen, L., Zhou, Y.L., Zhang, F.L., Shao, C.W., Zhang, X., Li, H.R.,
Wang, D., Zhang, D.E., and Fu, X.D. (2022)
Systematic Evaluation of Different R-loop Mapping Methods: Achieving Consensus,
Resolving Discrepancies and Uncovering Distinct Types of RNA:DNA Hybrids. (*co-first authors)
BioRxiv. DOI:10.1101/2022.02.18.480986

4. Lim, D.H.*#, Choi, M.S.*, Jeon, J.W.*, and Lee, Y.S.# (2023)
MicroRNA miR-252-5p regulates the Notch signaling pathway by targeting Rab6 in Drosophila
wing development. (*co-first authors / #co-corresponding authors)
Insect Science 30(5), 1431-1444 PMID: 36847222


5. Kim, C.J.*, Kim, H.H.*, Kim, H.K., Lee, S.J., Jang D.G., Kim, C.H., and Lim, D.H. (2023)
MicroRNA miR-263b-5p regulates developmental growth and cell association by suppressing
Laminin A in Drosophila.
Biology 12(8), 1096 (*co-first authors) PMID: 37626982

2024 ~

in Preparation (miR-1)
in Preparation (miR-33)
34. Lee, T.H.*, Kim, C.J.*, Lim, D.H.#, and Lee, Y.S.# (2025)
MicroRNA miR-315-5p regulates developmental growth in Drosophila wings by targeting S6k.
(*co-first authors / #co-corresponding authors)
Insect Science PMID: 40166978

33. Lim, B.H., Kim, S.C., Kim, H.J., Seo, Y.J., Lim, C.W., Park, Y.J., Sheet, S., Kim, D.H.,
Lim, D.H., Park, K.S., Lee, K.T., Kim, W.I., and Kim, J.M. (2025)
Single-cell transcriptomics of bronchoalveolar lavage during PRRSV infection with different
virulence.
Nature Communications 16(1):1112 PMID: 39875369

32. Kim, C.J.*, Jang, D.G.*, and Lim, D.H. (2024)
Drosophila miR-263b-5p controls wing developmental growth by targeting Akt
Animal Cells and Systems 29(1):35-45 PMID: 39777023

31. Jang, D.G.*, Kim, C.J.*, Shin, B.H.*, and Lim, D.H. (2024)
The biological roles of microRNAs in Drosophila development. (*co-first authors)
Insects 15(7):491 PMID: 39057224 [Review paper]


30. Jayne, N.D., Liang, Z, Lim, D.H., Chen, P.B., Diaz, C, Arimoto, K.-I., Xia, L., Ren, B., Fu, X.-D.,
and Zhang, D.-E. (2024)
RUNX1 C-terminal Mutations Impair Blood Cell Differentiation by Perturbing Specific Enhancer
-Promoter Networks.
Blood Advances 8(10):2410-2423 PMID: 38513139

2021 - 2023
29. Na, D.K*, Lim, D.H.*, Hong, J.S., Lee, H.M., Cho, D.A., Yu, M.S., Shaker, B., Ren, J.,
Lee, B.M., Song, J.G., Oh, Y.N., Lee, K.G., Oh, K.S., Lee, M.Y., Choi, M.S., Choi, H.S.,
Kim, Y.H., Bui, J.M., Leem K.S., Kim, H.W., Lee, Y.S.#, and Gsponer J.# (2023)
A multi-layered network model identifies Akt1 as a common modulator of neurodegeneration.
(*co-first authors)
Molecular Systems Biology e11801 PMID: 37984409

28. Kim, H.K., Kim, C.J., Jang, D.G., and Lim, D.H. (2023)
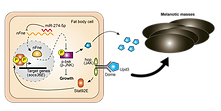
MicroRNA miR-274-5p suppresses found-in-neurons associated with melanotic mass
formation and development growth in Drosophila.
Insects 14(8), 709 PMID: 37623419
Preprints - doi: 10.20944/preprints202306.0501.v1
27. Kim, C.J.*, Kim, H.H.*, Kim, H.K., Lee, S.J., Jang D.G., Kim, C.H., and Lim, D.H. (2023)
MicroRNA miR-263b-5p regulates developmental growth and cell association by suppressing
Laminin A in Drosophila. (*co-first authors)
Biology 12(8), 1096 PMID: 37626982

26. Lee, S.J.*, Kim, N.Y.*, Jang, D.G., Kim, H.K., Kim, J.I., Jeon, J.W., and Lim, D.H. (2023)
Ecdysone-induced microRNA miR-276a-3p controls developmental growth by targeting the
insulin receptor in Drosophila. (*co-first authors)
Insect Molecular Biology 32(6):703-715 PMID: 37702106

25. Lim, D.H.*#, Choi, M.S.*, Jeon, J.W.*, and Lee, Y.S.# (2023)
MicroRNA miR-252-5p regulates the Notch signaling pathway by targeting Rab6 in Drosophila
wing development. (*co-first authors / #co-corresponding authors)
Insect Science 30(5), 1431-1444 PMID: 36847222


24. Jayne, N, Liu, M.D., Chen, B.S., Liang, Z.Y., Lim, D.H., Ren, B., Fu, X.D., Zhang, D.E. (2022)
Pathogenic RUNX1 C-Terminal Truncation Mutations Contribute to Differentiation Block and
Result in Unique Changes in Transcription and Chromatin Organization.
Blood 140(supplement 1), 996-997 DOI:10.1182/blood-2022-163838
23. Chen, J.Y.*, Lim, D.H.*, Chen, L., Zhou, Y.L., Zhang, F.L., Shao, C.W., Zhang, X., Li, H.R.,
Wang, D., Zhang, D.E., and Fu, X.D. (2022)
Systematic Evaluation of Different R-loop Mapping Methods: Achieving Consensus,
Resolving Discrepancies and Uncovering Distinct Types of RNA:DNA Hybrids. (*co-first authors)
BioRxiv. DOI:10.1101/2022.02.18.480986

22. Li, L.X., Luo, H.F., Lim, D.H., Han, L., Li, Y., Fu, X.D., and Qi, Y.J. (2021)
Global profiling of RNA-chromatin interactions reveals co-regulatory gene expression
networks in Arabidopsis.
Nature Plants 7(10), 1364-1378 PMID:34650265

2018 - 2020
21. Hao, Y.J., Wang, D.P., Wu, S.H., Li, X., Shao, C.W., Zhang, P., Chen, J.Y., Lim, D.H., Fu, X.D., He, S.M., and Chen,
R.S. (2020) Active Retrotransposons Help Maintain Pericentromeric Heterochromatin Required for Faithful Cell Division.
Genome Res. 30(11), 1570-1582. PMID:33060173
20. Lee, S.J., Hong, J.S., Lim, D.H., and Lee, Y.S. (2020) Roles for Drosophila Cap1 2’-O-ribose methyltransferase in the
small RNA silencing pathway associated with Argonaute 2. Insect Biochem. Mol. Biol. 123, 103415. PMID: 32504809
19. Lim, D.H.*, Lee, S.J.*, Chou, M.S., Han, J.Y., Seong, Y.M., Na, D.K., Kwon, Y.S., and Lee, Y.S. (2020) The conserved
microRNA miR-8-3p coordinates the expression of V-ATPase subunits to regulate ecdysone biosynthesis for Drosophila
metamorphosis. FASEB J. 34(5), 6449-6465 (*co-first authors). PMID: 32196731
18. Gou, L.T.*, Lim, D.H.*, Ma, W.B.*, Aubol, B.E., Hao, Y.J., Wang, X., Zhao, J., Liang, Z.Y., Shao, C.W., Zhang, X., Meng,
F., Li, H.R., Zhang, X.R., Xu, R.M., Li, D.S., Rosenfeld, M.G., Mellon, P.L., Adams, J.A., Liu, M.F., and Fu, X.D. (2020)
Initiation of Parental Genome Reprogramming in Fertilized Oocyte by Splicing Kinase SRPK1-Catalyzed Protamine
Phosphorylation. Cell 180(6), 1212-1227 (*co-first authors). PMID: 3216915
17. Chen, J.Y., Lim, D.H., and Fu, X.D. (2020) Mechanistic Dissection of RNA Binding Proteins in Regulated Gene
Expression at Chromatin Levels. Cold Spring Harb. Symp. Quant. Biol. 84, 55-66. PMID: 31900328
16. Zhou, B., Li, X., Luo, D., Lim, D.H., Zhou, Y., and Fu, X.D. (2019). GRID-seq for comprehensive analysis of global RNA-
chromatin interactions. Nature Protocols 14(7), 2036-2068. PMID: 31175345
15. Lim, D.H.*, Lee, S.*, Han, J.Y.*, Choi, M.S., Hong, J.S., and Lee, Y.S. (2019). MicroRNA miR-252 targets mbt to control
the developmental growth of Drosophila. Insect Mol. Biol. 28(3), 444-454. (*co-first authors). PMID: 30582233
14. Lim, D.H.*, Lee, S.*, Han, J.Y., Choi, M.S., Hong, J.S., Seong, Y., Kwon, Y.S., and Lee, Y.S. (2018). Ecdysone-
responsive microRNA-252-5p controls the cell cycle by targeting Abi in Drosophila. FASEB J. 32(8), 4519-4533. (*co-
first authors). PMID: 29543534
13. Bao, X., Guo, X., Yin, M., Tariq, M., Lai, Y., Kanwal, S., Zhou, J., Li, N., Lv, Y., Pulido-Quetglas, C., Wang, X., Ji, L.,
Khan, M.J., Zhu, X., Luo, Z., Shao, C., Lim, D.H., Liu, X., Li, N., Wang, W., He, M.H., Liu, Y.L., Ward, C., Wang, T. ,
Zhang, G., Wang, D.Y., Yang, J.H., Chen, Y.W., Zhang, C.L., Jauch, R., Yang, Y.G., Wang, Y.M., Qin, B.M., Anko, M.L.,
Hutchins, A.P., Sun, H., Wang, H.T., Fu, X.D., Zhang, B.L., and Esteban, M.A. (2018). Capturing the interactome of
newly transcribed RNA. Nature Methods 15(3), 213-220. PMID: 29431736
12. Lim, J.H., Kim, D.J., Lee, D.E., Han, J.Y., Chung, J.H., Ahn, H.K., Lee, S.W., Lim, D.H., Lee, Y.S., Park, S.Y., and Ryu,
H.M. (2015). Genome-wide microRNA expression profiling in placentas of fetuses with Down syndrome. Placenta.
36(3), 322-328.
11. Seong, Y., Lim, D.H., Kim, A., Seo, J.H., Lee, Y.S., Song, H., and Kwon, Y.S. (2014). Global identification of target
recognition and cleavage by the Microprocessor in human ES cells. Nucleic Acids Res. 42(20), 12806-12821.
10. Lim, D.H., Oh, C.T., Han, S.J., and Lee, Y.S. (2014). Methods for studying the biological consequences of endo-siRNA
deficiency in Drosophila melanogaster. Methods Mol. Biol. 1173, 51-58.
9. Joo, J.Y., Lee, J., Ko, H.Y., Lee, Y.S., Lim, D.H., Kim, E.Y., Cho, S., Hong, K.S., Ko, J.J., Lee, S., Lee, Y.S., Choi, Y.S.,
Lee, K.A., and Kim, S.H. (2014). Microinjection free delivery of miRNA inhibitor into zygotes. Sci. Rep. 4, 5417.
8. Lim, D.H., Lee, L., Oh, C.T., Kim, N.H., Hwang, S., Han, S.J., and Lee, Y.S. (2013). Microarray analysis of Drosophila
dicer-2 mutants reveals potential regulation of mitochondrial metabolism by endogenous siRNAs. J. Cell. Biochem.
114(2), 418-427.
7. Kwak, G.H., Lim, D.H., Han, J.Y., Lee, Y.S., and Kim, H.Y. (2012). Methionine sulfoxide reductase B3 protects from
endoplasmic reticulum stress in Drosophila and in mammalian cells. Biochem. Biophys. Res. Commun. 420(1), 130-
135.
6. Lim, D.H., Han, J.Y., Kim, J.R., Lee, Y.S., and Kim, H.Y. (2012). Methionine sulfoxide reductase B in the endoplasmic
reticulum is critical for stress resistance and aging in Drosophila. Biochem. Biophys. Res. Commun. 419(1), 20-26.
5. Lim, D.H.*, Oh, C.T.*, Lee, L., Hong, J.S., Noh, S.H., Hwang, S., Kim, S., Han, S.J., and Lee, Y.S. (2011). The
endogenous siRNA pathway in Drosophila impacts stress resistance and lifespan by regulating metabolic homeostasis.
FEBS Lett. 585(19), 3079-3085. (*co-first authors)
4. Cho, I.S., Kim, J., Seo, H.Y., Lim, D.H., Hong, J.S., Park, Y.H., Park, D.C., Hong, K.C., Whang, K.Y., and Lee, Y.S.
(2010). Cloning and characterization of microRNAs from porcine skeletal muscle and adipose tissue. Mol. Biol. Rep.
37(7), 3567-3574.
3. Lee, Y.S., Pressman, S., Andress, A.P., Kim, K., White, J.L., Cassidy, J.J., Li, X., Lubell, K., Lim, D.H., Cho, I.S.,
Nakahara, K., Preall, J.B., Bellare, P., Sontheimer, E.J., and Carthew, R.W. (2009). Silencing by small RNAs is linked to
endosomal trafficking. Nature Cell Biology 11(9), 1150-1156.
2. Cho, I.S., Kim, J., Lim, D.H., Ahn, H.C., Kim, H., Lee, K.B., and Lee, Y.S. (2008). Improved serum stability and
biophysical properties of siRNAs following chemical modifications. Biotechnol. Lett. 30(11), 1901-1908.
1. Lim, D.H.*, Kim, J.*, Kim, S.U., Carthew, R.W., and Lee, Y.S. (2008). Functional analysis of dicer-2 missense mutations in the siRNA pathway of Drosophila. Biochem. Biophys. Res. Commun. 371(3), 525-530. (*co-first authors)
2008-2015